Technical Advancements In Immunohistochemistry
Nov 27th 2019
There are a variety of techniques in molecular biology that can estimate protein expression levels or quantify your protein of interest. But how do we map the spatial arrangement of cells and protein targets at a particular physiological time point? Is there a technique that can read out protein expression, distribution, and activation in-situ?
Immunohistochemistry (IHC) is a relatively simple and cost-effective technique used to probe into the molecular morphology of cells and tissue micro-architecture. It uses the Antigen-Antibody interaction to visualize, analyze, quantify and phenotype cells at a tissue level. This technique can provide an insight into the tissue micro-environment and help understand the morphology, protein expression pattern and localization of various molecules. Hence, IHC can elucidate the complex intra, inter and extra-cellular interactions within a tissue matrix during a particular biological state. IHC is a well-established diagnostic tool to interpret disease heterogeneity in cancer and other diseases. It has been widely used to study the role of immune cells within solid tumors and their micro-environment.
The IHC technique has evolved over the last few years to improve the precision, accuracy and analytical features that can expedite research and enhance patient care.
The principle and applications of IHC
IHC is a technique in which specific antibodies (Ab) are used to detect a particular antigen (Ag)/protein within a cell or cells in a tissue, culture or smear. The Ab is labelled with a chromogenic or fluorescent tag to help visualization under a microscope. Most commonly, a polymer of labelled secondary Ab is used that can bind to the primary Ab, enhancing assay sensitivity and signal strength. A variation of immunohistochemistry, called in-situ hybridization (ISH), is similarly used to detect nuclear markers (DNA/RNA) using a probe complimentary to the genomic sequence.
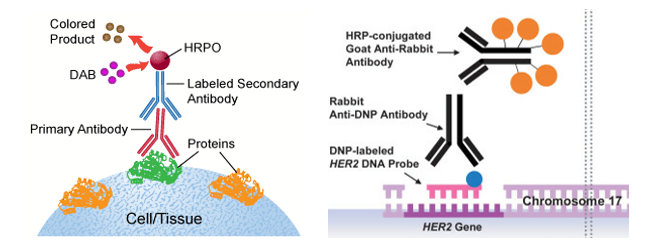
Figure 1: (A) The principle of 2-step IHC. The 2 o Ab is labelled with horse-radish peroxidase (HRP) enzyme which gives a brown precipitate when it reacts with DAB. (B) The principle of in-situ hybridization. A labelled probe, complimentary to the genomic DNA sequence, is detected by a primary and labelled secondary Ab giving a signal.
The applications of immunohistochemistry spans clinical, translational, and basic research. IHC is a very common clinical diagnostic tool used for prediction of treatment, prognosis and outcome. IHC can help identify abnormal cells, pathological conditions or disease stages by identifying biomarkers that are specific to a certain cell type, cellular phase or infection. Moreover, IHC has been widely used in drug development to identify and validate pharmaceutical targets, analyze tumor-stroma-immune cell interactions and stratify patients into progression-risk categories in clinical trials.
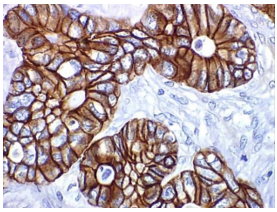
In recent times, IHC has been most extensively implemented in oncology research to investigate the cancer proteome. The prime focus has been on evaluating the cellular context pertaining to the different stages of tumorigenesis, biological crosstalks involved, identification of potential therapeutic targets, as well as screening prognostic or predictive biomarkers.
To cope with the increasing demand for accurate and precise data, the technique of IHC has undergone rapid transformative development, leading to the advent of sophisticated modifications that can capture more contextual information.
Figure 2: The IHC staining showing membrane and cytoplasmic positivity for a particular biomarker in colon cancer cells. The brown color is the chromogen DAB showing the presence and localisation of target protein. The blue is the hematoxylin counterstain that stains the cell nucleus.
Multiplex IHC
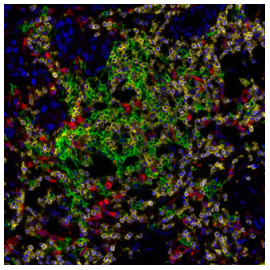
With an expansion in immunotherapeutic research and the need for profiling tumor micro-environments, the conventional IHC technique has been tweaked at different levels for more comprehensive results. Conventional IHC uses serial section from the same sample to analyze different proteins or biomarkers. This approach can exhaust the sample and also affect assay reproducibility. On the other hand, multiplex IHC or multiple immune-labelling can maximize the amount of information derived from a single or limited sample. Multiplexing can effectively elucidate cellular interactions and co-localization of proteins.
Chromogenic IHC can be multiplexed only up to dual or triple markers with good spatial resolution, whereas fluorescent detection offers a higher degree of multiplexing. This is usually achieved by stain cycling that involves repeated cycles of antibody staining, imaging and quenching. Tyramide signal amplification (TSA), MultiOmyx TM and fluorescent quantum dot nano-crystals are some of the latest multiplex technologies that enables detection of low abundance targets, as well as enhances the signal to noise ratio.
Figure 3: Detection of human CD3 (yellow), CD20 (green), and CD68 (red) in FFPE lung carcinoma by IHC-IF. Reference: www.biomol.com
However, multiplex IHC/IF does have its limitations. Spectral overlap of multiple dyes or color mixing in co-localized proteins, and tissue auto-fluorescence can make it difficult to interpret results. Besides, stain cycling methods have cumbersome protocols with low throughput and quality assurance can be debatable. Some of the latest innovations combining IHC and mass spectroscopy have shown to overcome these limitations enabling high order multiplexing and analysis.
Moreover, an important outcome of multiplex IHC has been the evolution of digital pathology and image analysis, creating a paradigm shift in tissue analysis approach.
Computational Pathology
This is the age of data driven medicine. Automation and machine learning concepts has been implemented in the field of IHC and pathology as well. Whole slide imaging and digital analysis of slides have been around for more than a decade. With the boom in cancer research and the growing need for precision medicine, there has been a rapid growth in the area of computational pathology. More so, the field is headed towards artificial intelligence. Digitization also permits remote analysis and addresses the problems of subjective bias accompanied with manual scoring.
Digital pathology and image analysis have important roles across drug development and companion diagnostics. Whole slide scanning followed by digital image analysis has truly revolutionized bio-banking, molecular pathology, tissue microarray analysis and molecular profiling of tissue. Different slide scanners are available in the market that scan and store whole slide images in various formats. Leica, Hamamatsu, Olympus, Philips, Roche and Zeiss are some of the prominent players in this arena. These images can be archived on cloud locations facilitating remote accessibility and ‘telepathology’. Furthermore, several image analysis software tools have been developed that can identify cells and tissue compartments giving cell counts and staining intensity. Some of the currently used tools include Halo by Indica labs, Definiens (recently acquired by MedImmune), PerkinElmer and Visiopharm.
However, such tools are being more widely used in pharmaceutical research rather than clinical laboratories. Some of the major applications include the study of tumor margins, tumor infiltrating lymphocytes (TILs) and the evaluation of biomarkers that correlate with drug response or survival.
The integration of artificial intelligence (AI) and data analysis has been the latest approach. Machine learning algorithms are being designed to automatically classify tissue compartments, segment cell types or anatomical features and make calculations based on biomarker expression levels, localization and proximity. Many developments are still underway to achieve fully automated, comprehensive and reliable solutions that can completely eliminate human bias and subjective errors to accelerate research and discovery.
Next-Generation IHC
When mass spectrometry (MS) is combined with IHC, the multiplexing ability can be increased several folds. The first MS-IHC techniques were described by two independent research groups, who came up with two slightly different approaches, scanning mass cytometry (SMC) and multiplexed ion-beam imaging (MIBI). Both these methods use antibodies labelled with isotopically pure metal tags, usually rare earth metals (lanthanides/actinides). This approach gives completely resolvable peaks derived from mass measurements of the metals creating zero spectral overlap. The two methods differ in how these tags are liberated from the sample, ionized, and detected. MS-IHC could be the new standard in pathology, allowing detection of the presence, abundance, location, and functional state of up to 100 targets giving an analytical read out from subcellular level up to whole organs.
Scanning Mass Cytometry
Developed by Giensen
et al. in 2014, SMC is an extension of CyTOF in which
laser ablation of tissue is coupled with plasma time-of-flight mass
spectrometry. A high-intensity laser beam of ~1 uM spot size is used to
liberate tissue into a carrier gas which is then transports the ablated sample
into an argon plasma where it is ionized and detected. An analysed 2-D image is
constructed using a software which looks very similar to a routine IHC but with
quantitative multiplexed information.
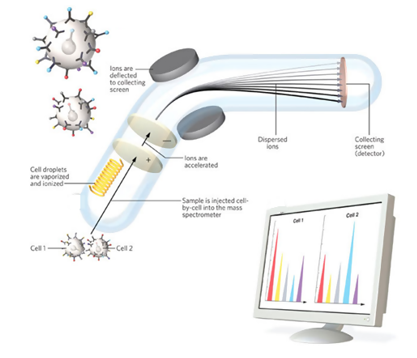
Figure 4: Principle and workflow of SMC. 1) Tissues are probed with Antibodies labelled with metal ions. 2) Laser beam releases the cells from tissue into an argon plasma which vaporizes the cell and strips electrons from the metal to form ions. The metal ions are accelerated in an electrostatic field and a deflector separates them depending on the mass. Mass spectrometer can precisely distinguish isotopes that differ by a single atomic mass unit allowing it to measure 100 targets in a single experiment. 3)The detector counts the arriving ions, quantifying the target.
Multiplexed Ion-Beam Imaging
MIBI, first described by Garry Nolan’s group, is based on secondary ion mass spectrometry (Angelo et al., 2014). This method uses a scanning ion beam of sun-micron spot size to scan the sample and liberate the ions which are detected using a magnetic sector mass spectrometer. Here, very tiny spots are vaporised at a time resulting in a higher resolution. However, MIBI requires more a specialized setup including vacuum and multiple detector MS. An imaging software finally creates the 2-D stained tissue image from the detected heavy metal ions. On the downside, MIBI currently uses magnetic sector detection, which permits only seven targets to be detected per scan; the mass sensors have to be re-positioned to detect new targets for each additional scan. (RM Levenson et al., 2015)
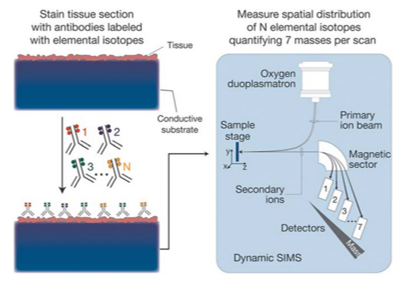
Figure 5: MIBI sample preparation and scanning procedures. Biological specimens, such as FFPE tissue or cell suspensions, are immobilized on a conductive substrate, such as indium tin oxide-coated glass or silicon wafer. Samples are subsequently stained with antibodies conjugated to unique transition element isotope reporters, dried, and loaded under vacuum for MIBI analysis. The sample surface is rasterized with a primary ion beam (O) that sputters the antibody-specific isotope reporters present on the sample surface as secondary ions. Metal-conjugated antibodies are quantified via replicate scans of the same field of view, during which up to seven metals reporters are measured with each scan (Referece: RM Levenson et al., 2015)
MIBI is considered to be more sensitive than SMC as the ionization yield is 1-10% compared to 0.01% in SMC. The resolution is also greater in MIBI which is 200 nm. Moreover, SMC is destructive and vaporizes the entire tissue sample whereas MIBI only consumes 5–10 nm of the top layer of the sample, allowing replicate scanning.
Advantages of MS-IHC
Metal tagged antibodies possess higher sensitivity, low crosstalk and high dynamic range which minimizes the challenges for validation even with high order multiplexing. A single step Ab-labeling protocol and digital imaging increases convenience and may reduce inter-laboratory variability. Also, the use of multiple monoclonal antibodies for the same protein can give some valuable insights on protein structure and post-translational modifications. 3D imaging and computations have also been proposed using MS-IHC.
Having said that, MS-IHC is still very naïve and further development is on-going to improve robustness, analytical outputs and cost-effectiveness. This cutting-edge technology has definitely opened the doors to high-dimensional capabilities that can put drug development research on the fast-track.
Future perspectives
The technical advancements in immunohistochemistry has the potential to unlock the immense wealth of information in tissue samples. In this era of personalized medicine, it enables researchers and pathologists to consistently deliver a higher quality of care.
With digital pathology and computational biology being the norm, stringent control and validation methods are required to streamline operations. Artificial intelligence can further leverage the way data is interpreted and analysed. Integration of epidemiological, clinical and genomic data can help reveal the deeper relationships between genotype and phenotype, driving individualized molecular diagnosis and therapies.
All these new innovations ultimately focus on delineating the molecular configurations of complex biological systems. Multiplexing, automation and multi-parametric analysis can bring new vistas in basic and translational science.
------
References
